Evolutionary origins of SARS-Cov-2 identified
Published: 30 July 2020
The Centre for Virus Research's Professor David Robertson has contributed towards research that has discovered the lineage that gave rise to the virus has been circulating in bats for decades and likely includes other viruses with the ability to infect humans.
The Centre for Virus Research's Professor David Robertson has contributed towards research that has discovered the lineage that gave rise to SARS-CoV-2 has been circulating in bats for decades and likely includes other viruses with the ability to infect humans
An international research team of Chinese, European and U.S. scientists made the discovery by reconstructing the evolutionary history of the virus responsible for the COVID-19 pandemic.
The findings, which were recently published in Nature Microbiology, could have implications for the prevention of future pandemics stemming from this lineage
Maciej Boni, Associate Professor of Biology at Penn State, said: "Coronaviruses have genetic material that is highly recombinant, meaning different regions of the virus’s genome can be derived from multiple sources. This has made it difficult to reconstruct SARS-CoV-2’s origins.
"You have to identify all the regions that have been recombining and trace their histories. To do that, we put together a diverse team with expertise in recombination, phylogenetic dating, virus sampling, and molecular and viral evolution.”
The team used three different bioinformatic approaches to identify and remove the recombinant regions within the SARS-CoV-2 genome.
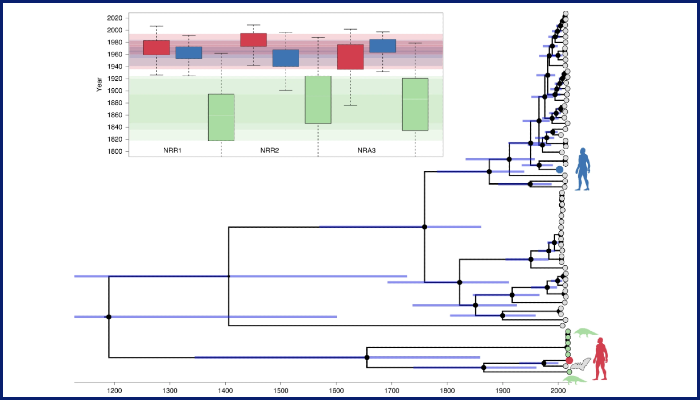
Next, they reconstructed phylogenetic histories for the non-recombinant regions and compared them to each other to see which specific viruses have been involved in recombination events in the past.
They were able to reconstruct the evolutionary relationships between SARS-CoV-2 and its closest known bat and pangolin viruses.
The researchers found that the lineage of viruses to which SARS-CoV-2 belongs diverged from other bat viruses about 40 to 70 years ago.
Although SARS-CoV-2 is genetically similar - about 96 per cent - to the RaTG13 coronavirus, which was sampled from a Rhinolophus affinis horseshoe bat in 2013 in Yunnan province, China, the team found that it diverged from RaTG13 a relatively long time ago, in 1969.
Philippe Lemey, Principal Investigator in the Department of Evolutionary and Computational Virology, KE Leuven, explained: "The ability to estimate divergence times after disentangling recombination histories, which is something we developed in this collaboration, may lead to insights into the origins of many different viral pathogens."
The team found that one of the older traits that SARS-CoV-2 shares with its relatives is the receptor-binding domain (RBD) located on the Spike protein, which enables the virus to recognize and bind to receptors on the surfaces of human cells.
David Robertson, Professor of Computational Virology at the CVR, added: "This means that other viruses that are capable of infecting humans are circulating in horseshoe bats in China."
Will these viruses be capable of jumping directly from bats into humans or will an intermediate species be required to make the leap?
According to Robertson, for SARS-CoV-2, other research groups incorrectly proposed that key evolutionary changes occurred in pangolins.
He said: “SARS-CoV-2’s RBD sequence has so far only been found in a few pangolin viruses. Furthermore, the other key feature thought to be instrumental to SARS-CoV-2’s ability to infect humans — a polybasic cleavage site insertion in the Spike protein — has not yet been seen in another close bat relative of the SARS-CoV-2 virus.
"Yet, while it is possible that pangolins may have acted as an intermediate host facilitating transmission of SARS-CoV-2 to humans, no evidence exists to suggest that pangolin infection is a requirement for bat viruses to cross into humans.
"Instead, our research suggests that SARS-CoV-2 likely evolved the ability to replicate in the upper respiratory tract of both humans and pangolins.”
The team concluded that preventing future pandemics will require better sampling within wild bats and the implementation of human disease surveillance systems that are able to identify novel pathogens in humans and respond in real time.
Robertson noted: "The key to successful surveillance is knowing what viruses to look for and prioritizing those that can readily infect humans. We should have been better prepared for a second SARS virus."
Boni added: "We were too late in responding to the initial SARS-CoV-2 outbreak - but this will not be our last coronavirus pandemic.
"A much more comprehensive and real-time surveillance system needs to be put in place to catch viruses like this when case numbers are still in the double digits.”
Support for this research was provided by the European Research Council, the Medical Research Council, the Research Foundation — Flanders and the National Natural Science Foundation of China.

Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic
- Maciej F. Boni, Philippe Lemey, Xiaowei Jiang, Tommy Tsan-Yuk Lam, Blair W. Perry, Todd A. Castoe, Andrew Rambaut & David L. Robertson. Nat Microbiol (2020).
Support for this research was provided by the European Research Council, the Medical Research Council, the Research Foundation — Flanders, and the National Natural Science Foundation of China.
Enquiries: ali.howard@glasgow.ac.uk or elizabeth.mcmeekin@glasgow.ac.uk / 0141 330 6557 or 0141 330 4831
First published: 30 July 2020

